Security: Model Registration and Approval
Federated learning in Fed-BioMed is performed by sending models to each node and by requesting nodes to train model. Hence, the content of model files plays an important role in terms of privacy and security. Therefore, Fed-BioMed offers a feature to run only pre-approved models. Model files that are sent by a researcher with the training request should be approved by the node side. This model approval process avoids possible attacks that come through the model code files, run into nodes, that may access private data or jeopardize the node. The approval process should be done by a real user/person who will review the model file. The reviewer should make sure the model doesn't contain any code that might cause privacy issues.
How the Models get Approved and Controlled
Model approval can be done through the Fed-BioMed CLI (Command Line Interface) before or after the node is started. Model files should be prepared as txt files. Each model should have a unique name and unique file path. During the registration, the ModelManager of the Fed-BioMed hashes the content of the model file and saves the hash into the persistent database (Approved Models in Figure 1). Model files that are sent by a researcher are first hashed and then compared to the saved hashes for approval. This process flow is presented in the following figure.
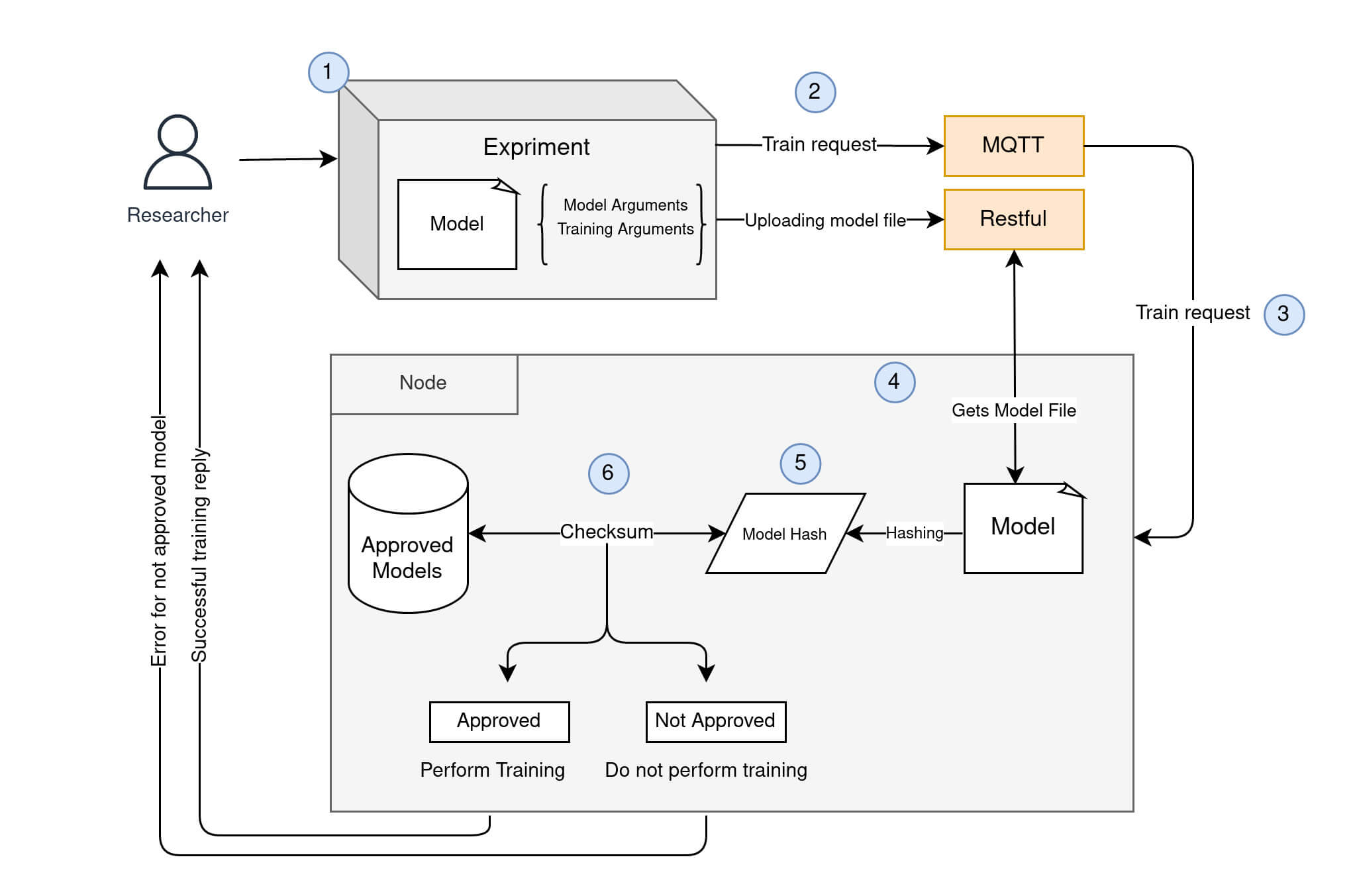
Details of Figure 1 workflow:
Researchercreates aModelto be trained on theNodeResearchersubmits aModelto theNode: he/she sends a train request to the node ...- ... and sends a model file containing the
Model NoderecievedModelsent byResearcherNodecomputes hash of the incomingModel.Nodechecks ifResearcher's model has already been approved by comparing its hash to the existing pool of approved models hashes.- If
Modelhas been approved by theNode,Researcherwill be able to train his/her model on theNode.
Hashing
ModelManager of Fed-BioMed provides various hashing algorithms. These hashing algorithms are guaranteed by the Python hashlib built-in library. The hashing algorithm can be selected by configuring the configuration file of the node. Provided hasing algorithms by Fed-BioMed are: SHA256, SHA384, SHA512, SHA3_256, SHA3_384, SHA3_512, BLAKE2B, and BLAKE2S. You can have more information about these hashing algorithms in hashlib documentation page.
Checksum Operation
Checksum operation is done by querying database for hash of the requested model. Therefore, the model files that are approved should be registered/saved into database by using Fed-BioMed CLI before the checksum operation (train request). This operation controls whether any registered/approved models matches the model requested.
ModelManager minifies model files just before hashing the model file. The minification process removes spaces and comments from the model file. The purpose of using minified models is to avoid errors when the requested model file has more or less comments or empty spaces than the model which is approved. Since the spaces and the comments will have no effect when executing models, this process will not open a back door for attacks. Therefore, having more spaces or comments than the registered model will not affect the checksum result (and thus the hashing).
Managing Nodes for Model Approval
Model approval can be managed either through configuration file or Fed-BioMed CLI. The configuration file of the node includes a section named security to control/switch options for selecting hashing algorithm, enabling/disabling model approval, and accepting default models as approved. By default, Fed-BioMed does not enables model approval. It means when you start or add data to the node for the first time, if the configuration file doesn't exist, it creates a new configuration file as model approval is disabled; model_approval = False.
Default Models
Default models are the model files that are created for Fed-BioMed tutorials, i.e. models contained in /notebooks folder. These models are saved into envs/development/default_models/ directory. If the node is configured to allow default models for training, it registers default models when the node is started. These models are saved for testing purposes, and they can be disabled in a production environment.
The hashes of the default model won't be updated while starting the node if the node is configured not to allow default models. However, default models might be already saved into database previously. Even if there are default models in the database, the node will not approve requests for the default models as long as this option has been disabled.
Config Files
When the new node is created without any specified configuration file or any options, the default configuration file is saved into the etc directory of Fed-BioMed as follows.
[default]
# other paramters
[mqtt]
# parameters for mqtt
[security]
hashing_algorithm = SHA256
allow_default_models = True
model_approval = False
model_approval is disabled. For enabling or disabling this feature, you can change its value to True or False. Any values different from True or False will be counted as False. The node should be restarted to apply changes after updating the config file. When the model approval is False, allow_default_models will have no effect because there will be no model control operation for train requests.
Changing Hashing Algorithm
By default, Fed-BioMed uses the SHA256 hashing algorithm to hash model files both for registering and checking. It can be changed based on provided algorithms by Fed-BioMed. These algorithms are already presented in the "Hashing" section of this article. After the hashing algorithm is changed, the node should be restarted. When restarting the node, if the model approval is enabled CLI will update hashes in the database by recreating them with the chosen hashing algorithm in the config file.
Using Fed-BioMed CLI
Fed-BioMed CLI has been configured to start nodes with certain parameters to managing model approval features. It is possible to change the default parameters of config file while starting a node for the first time. For instance, the following command will disable model approval and allow default models for the node. Let's assume we are working with a config file called config-n1.ini. If the config-n1.ini file doesn't exist, it will create the config-n1.ini file with the parameters model_approval = False and allow_default_models = False, under [security] sub-section.
$ {FEDBIOMED_DIR}/scripts/fedbiomed_run node config config-n1.ini --disable-model-approval --disable-default-models start
It is also possible to start node by enabling model_approval mode, even it is disabled in the configuration file. For instance, suppose that the config-n1.ini file is saved as follows,
[security]
hashing_algorithm = SHA256
allow_default_models = False
model_approval = False
The command below will force the node to start it with enabled model approval mode.
$ {FEDBIOMED_DIR}/scripts/fedbiomed_run node config config-n1.ini --enable-model-approval --allow-default-models start
$ {FEDBIOMED_DIR}/scripts/fedbiomed_run node config config-n1.ini --enable-model-approval --disable-default-models start
Hashing algorithm should be changed directly from the configuration file.
Model Registration
Currently, the model registration process is done by the Fed-BioMed CLI tool. Model files that will be registered should be stored in the file system as txt files.
Node: Registering New Model
The following command will launch Fed-BioMed CLI for selecting a model file and entering a name and description for the model. The model name and of course its path should be unique. It means that you can not add same model file multiple times.
$ {FEDBIOMED_DIR}/scripts/fedbiomed_run node config config-n1.ini register-model
ModelManager will create has for the model file and save it into a persistent database with following details. 'name' : '<model-name>'
'description' : '<description>'
'path' : '<path/to/model/fıle>'
'hash' : '<hash of the model file>'
'date_registered' : '<Registeration date>'
'date_created' : '<The date file has been created>'
'date_modified' : '<The date file has been modified>'
'model_type' : 'registered'
model_type will be always registered for the models that are saved through Fed-BioMed CLI. This information indicates that the model isn't a default model provided by Fed-BioMed.
Each time when the node is started, ModelManager checks whether the model file is deleted from the file system. If it is deleted, ModelManager also deletes it from the database. Therefore, please make sure that the model file is always exists in the path where it is stored.
Deleting Models
Following command is used for deleting registered models.
$ {FEDBIOMED_DIR}/scripts/fedbiomed_run node config config-n1.ini delete-model
Select the model to delete:
1) MyModel Model ID model_98a1e68d-7938-4889-bc46-357e4ce8b6b5
2) MyModel2 Model ID model_18314625-2134-3334-vb35-123f3vbe7fu7
Select:
Updating Models
It is possible update registered models with different or same model file that has different content. This is usefull when working on a model and you want it to be updated without having to remove it and restore it in database. Following command launches the CLI to select the model that will be updated
$ {FEDBIOMED_DIR}/scripts/fedbiomed_run node config config-n1.ini update-model
You can update hashes only by providing a model file. This API does not allow you to update saved hashes directly.
Default Models
Unlike the registered model, the Fed-BioMed CLI tool doesn't provide a option for adding new default models. Default models are already stored in the envs/development/default_models directory, and they are automatically registered when the node is started with model_type as default. If the default models have already been added to the database, ModelManager checks whether there is any modification. If any default model file is deleted, ModelManager also deletes them from the database. If the model file is modified, or the hashing algorithm is changed, ModelManager updates hashes. This checking/controlling operation is done while starting the node.
Default models can not be removed using Fed-BioMed CLI. They should be removed from the envs/development/default_models directory. After restarting the node, deleted model files will be also removed from the Models table of the node database.


