MNIST classification with Scikit-Learn Classifier (Perceptron)¶
Overview of the tutorial:
In this tutorial, we are going to train Scikit-Learn Perceptron as a federated model model over a Node.
At the end of this tutorial, you will learn:
- how to define a Sklearn classifier in Fed-BioMed (especially
Perceptronmodel) - how to train it
- how to evaluate the resulting model
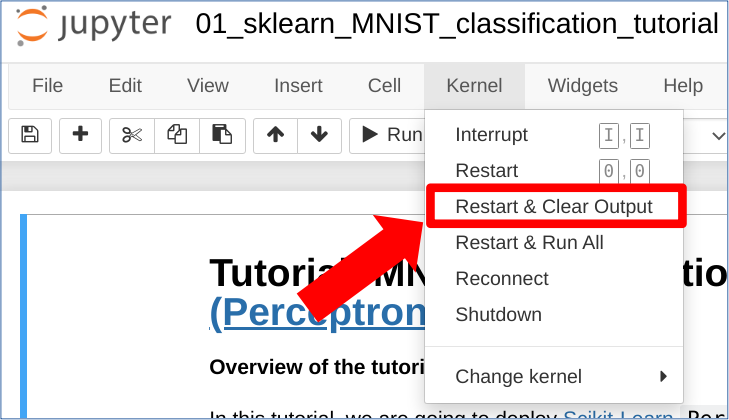
HINT : to reload the notebook, please click on the following button:
Kernel -> Restart and clear Output
1. Clean your environments¶
Before executing notebook and starting nodes, it is safer to remove all configuration scripts automatically generated by Fed-BioMed. To do so, enter the following in a terminal:
source ${FEDBIOMED_DIR}/scripts/fedbiomed_environment cleanNote: ${FEDBIOMED_DIR} is a path relative to based directory of the cloned Fed-BioMed repository. You can set it by running command export FEDBIOMED_DIR=/path/to/fedbiomed. This is not required for Fed-BioMed to work but enables you to run the tutorials more easily.
2. Setting the node up¶
It is necessary to previously configure a Network and a Nodebefore runnig this notebook:
${FEDBIOMED_DIR}/scripts/fedbiomed_run network${FEDBIOMED_DIR}/scripts/fedbiomed_run node add- Select option 2 (default) to add MNIST to the node
- Confirm default tags by hitting "y" and ENTER
- Pick the folder where MNIST is downloaded (this is due torch issue https://github.com/pytorch/vision/issues/3549)
- Data must have been added (if you get a warning saying that data must be unique is because it has been already added)
Check that your data has been added by executing
${FEDBIOMED_DIR}/scripts/fedbiomed_run node list- Run the node using
${FEDBIOMED_DIR}/scripts/fedbiomed_run node start. Wait until you getConnected with result code 0. it means your node is working and ready to participate to a Federated training.
More details are given in tutorial : Installation/setting up environment
3. Create Sklearn Federated Perceptron Training Plan¶
The class FedPerceptron constitutes the Fed-BioMed wrapper for executing Federated Learning using Scikit-Learn Perceptron model based on mini-batch Stochastic Gradient Descent (SGD). As we have done with Pytorch model in previous chapter, we create a new training plan class SkLearnClassifierTrainingPlan that inherits from it. For a refresher on how Training Plans work in Fed-BioMed, please refer to our Training Plan user guide.
In scikit-learn Training Plans, you typically need to define only the training_data function, and optionally an init_dependencies function if your code requires additional module imports.
The training_data function defines how datasets should be loaded in nodes to make them ready for training. It takes a batch_size argument and returns a DataManager class. For scikit-learn, the DataManager must be instantiated with a dataset and a target argument, both np.ndarrays of the same length.
from fedbiomed.common.training_plans import FedPerceptron
from fedbiomed.common.data import DataManager
import numpy as np
class SkLearnClassifierTrainingPlan(FedPerceptron):
def init_dependencies(self):
"""Define additional dependencies.
return ["from torchvision import datasets, transforms",
"from torch.utils.data import DataLoader"]
def training_data(self, batch_size):
In this case, we rely on torchvision functions for preprocessing the images.
"""
return ["from torchvision import datasets, transforms",]
def training_data(self, batch_size):
"""Prepare data for training.
This function loads a MNIST dataset from the node's filesystem, applies some
preprocessing and converts the full dataset to a numpy array.
Finally, it returns a DataManager created with these numpy arrays.
"""
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
dataset = datasets.MNIST(self.dataset_path, train=True, download=False, transform=transform)
X_train = dataset.data.numpy()
X_train = X_train.reshape(-1, 28*28)
Y_train = dataset.targets.numpy()
return DataManager(dataset=X_train, target=Y_train, batch_size=batch_size, shuffle=False)
Provide dynamic arguments for the model and training. These may potentially be changed at every round.
Model arguments¶
model_args is a dictionary with the arguments related to the model, that will be passed to the Perceptron constructor.
IMPORTANT For classification tasks, you are required to specify the following two fields:
n_features: the number of features in each input sample (in our case, the number of pixels in the images)n_classes: the number of classes in the target data
Furthermore, the classes may not be represented by arbitrary values: classes must be identified by integers in the range 0..n_classes
Training arguments¶
training_args is a dictionary containing the arguments for the training routine (e.g. batch size, learning rate, epochs, etc.). This will be passed to the routine on the node side.
model_args = {'n_features': 28*28,
'n_classes' : 10,
'eta0':1e-6,
'random_state':1234,
'alpha':0.1 }
training_args = {
'epochs': 3,
'batch_maxnum': 20, # can be used to debugging to limit the number of batches per epoch
# 'log_interval': 1, # output a logging message every log_interval batches
'batch_size': 4
}
4. Train your model on MNIST dataset¶
MNIST dataset is composed of handwritten digits images, from 0 to 9. The purpose of our classifier is to associate an image to the corresponding represented digit
from fedbiomed.researcher.experiment import Experiment
from fedbiomed.researcher.aggregators.fedavg import FedAverage
tags = ['#MNIST', '#dataset']
rounds = 3
# select nodes participating in this experiment
exp = Experiment(tags=tags,
model_args=model_args,
training_plan_class=SkLearnClassifierTrainingPlan,
training_args=training_args,
round_limit=rounds,
aggregator=FedAverage(),
node_selection_strategy=None)
exp.run(increase=True)
5. Testing on MNIST test dataset¶
Let's assess performance of our classifier with MNIST testing dataset
import tempfile
import os
from fedbiomed.researcher.environ import environ
from torchvision import datasets, transforms
from sklearn.preprocessing import LabelBinarizer
import numpy as np
tmp_dir_model = tempfile.TemporaryDirectory(dir=environ['TMP_DIR']+os.sep)
model_file = os.path.join(tmp_dir_model.name, 'class_export_mnist.py')
# collecting MNIST testing dataset: for that we are downloading the whole dataset on en temporary file
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
testing_MNIST_dataset = datasets.MNIST(root = os.path.join(environ['TMP_DIR'], 'local_mnist.tmp'),
download = True,
train = False,
transform = transform)
testing_MNIST_data = testing_MNIST_dataset.data.numpy().reshape(-1, 28*28)
testing_MNIST_targets = testing_MNIST_dataset.targets.numpy()
6. Getting Loss function¶
Here we use the aggregated_params() getter to access all model weights at the end of each round to plot the evolution of Percpetron loss funciton, as well as its accuracy.
# retrieve Sklearn model and losses at the end of each round
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import accuracy_score, confusion_matrix, hinge_loss
fed_perceptron_model = exp.training_plan().model()
perceptron_args = {key: model_args[key] for key in model_args.keys() if key in fed_perceptron_model.get_params().keys()}
losses = []
accuracies = []
for r in range(rounds):
fed_perceptron_model = fed_perceptron_model.set_params(**perceptron_args)
fed_perceptron_model.classes_ = np.unique(testing_MNIST_dataset.targets.numpy())
fed_perceptron_model.coef_ = exp.aggregated_params()[r]['params']['coef_'].copy()
fed_perceptron_model.intercept_ = exp.aggregated_params()[r]['params']['intercept_'].copy()
prediction = fed_perceptron_model.decision_function(testing_MNIST_data)
losses.append(hinge_loss(testing_MNIST_targets, prediction))
accuracies.append(fed_perceptron_model.score(testing_MNIST_data,
testing_MNIST_targets))
7. Comparison with a local Perceptron model¶
In this section, we implement a local Perceptron model, so we can compare remote and local models accuracies.
You can use this section as an insight on how things are implemented within the Fed-BioMed network. In particular, looking at the code in the next few cells you may learn how:
- we implement mini-batch gradient descent for scikit-learn models
- we implement
Perceptronbased onSGDClassifier
# downloading MNIST dataset
training_MNIST_dataset = datasets.MNIST(root = os.path.join(environ['TMP_DIR'], 'local_mnist.tmp'),
download = True,
train = True,
transform = transform)
training_MNIST_data = training_MNIST_dataset.data.numpy().reshape(-1, 28*28)
training_MNIST_targets = training_MNIST_dataset.targets.numpy()
Local Model training loop : a new model is trained locally, then compared with the remote FedPerceptron model
fed_perceptron_model.get_params()
local_perceptron_losses = []
local_perceptron_accuracies = []
classes = np.unique(training_MNIST_targets)
batch_size = training_args["batch_size"]
# model definition
local_perceptron_model = SGDClassifier()
perceptron_args = {key: model_args[key] for key in model_args.keys() if key in fed_perceptron_model.get_params().keys()}
local_perceptron_model.set_params(**perceptron_args)
model_param_list = ['coef_', 'intercept_']
# Model initialization
local_perceptron_model.intercept_ = np.zeros((model_args["n_classes"],))
local_perceptron_model.coef_ = np.zeros((model_args["n_classes"], model_args["n_features"]))
Implementation of mini-batch SGD
for r in range(rounds):
for e in range(training_args["epochs"]):
tot_samples_processed = 0
for idx_batch in range(training_args["batch_maxnum"]):
param = {k: getattr(local_perceptron_model, k) for k in model_param_list}
grads = {k: np.zeros_like(v) for k, v in param.items()}
# for each sample: 1) call partial_fit 2) accumulate the gradients 3) reset the model parameters
for sample_idx in range(tot_samples_processed, tot_samples_processed+batch_size):
local_perceptron_model.partial_fit(training_MNIST_data[sample_idx:sample_idx+1,:],
training_MNIST_targets[sample_idx:sample_idx+1],
classes=classes)
for key in model_param_list:
grads[key] += getattr(local_perceptron_model, key)
setattr(local_perceptron_model, key, param[key])
tot_samples_processed += batch_size
# after each epoch, we update the model with the averaged gradients over the batch
for key in model_param_list:
setattr(local_perceptron_model, key, grads[key] / batch_size)
predictions = local_perceptron_model.decision_function(testing_MNIST_data)
local_perceptron_losses.append(hinge_loss(testing_MNIST_targets, predictions))
local_perceptron_accuracies.append(local_perceptron_model.score(testing_MNIST_data,
testing_MNIST_targets))
Compare the local and federated models. The two curves should overlap almost identically, although slight numerical errors are acceptable.
import matplotlib.pyplot as plt
plt.figure(figsize=(10,5))
plt.subplot(1,2,1)
plt.plot(losses, label="federated Perceptron losses")
plt.plot(local_perceptron_losses, "--", color='r', label="local Perceptron losses")
plt.ylabel('Perceptron Cost Function (Hinge)')
plt.xlabel('Number of Rounds')
plt.title('Perceptron loss evolution on test dataset')
plt.legend()
plt.subplot(1,2,2)
plt.plot(accuracies, label="federated Perceptron accuracies")
plt.plot(local_perceptron_accuracies, "--", color='r',
label="local Perceptron accuracies")
plt.ylabel('Accuracy')
plt.xlabel('Number of Rounds')
plt.title('Perceptron accuracy over rounds (on test dataset)')
plt.legend()
In this example, plots appear to be the same: this means that Federated and local Perceptron models are performing equivalently!
8. Getting accuracy and confusion matrix¶
# federated model predictions
fed_prediction = fed_perceptron_model.predict(testing_MNIST_data)
acc = accuracy_score(testing_MNIST_targets, fed_prediction)
print('Federated Perceptron Model accuracy :', acc)
# local model predictions
local_prediction = local_perceptron_model.predict(testing_MNIST_data)
acc = accuracy_score(testing_MNIST_targets, local_prediction)
print('Local Perceptron Model accuracy :', acc)
def plot_confusion_matrix(fig, ax, conf_matrix, title, xlabel, ylabel, n_image=0):
im = ax[n_image].imshow(conf_matrix)
ax[n_image].set_xticks(np.arange(10))
ax[n_image].set_yticks(np.arange(10))
for i in range(conf_matrix.shape[0]):
for j in range(conf_matrix.shape[1]):
text = ax[n_image].text(j, i, conf_matrix[i, j],
ha="center", va="center", color="w")
ax[n_image].set_xlabel(xlabel)
ax[n_image].set_ylabel(ylabel)
ax[n_image].set_title(title)
fed_conf_matrix = confusion_matrix(testing_MNIST_targets, fed_prediction)
local_conf_matrix = confusion_matrix(testing_MNIST_targets, local_prediction)
fig, axs = plt.subplots(nrows=1, ncols=2,figsize=(10,5))
plot_confusion_matrix(fig, axs, fed_conf_matrix,
"Federated Perceptron Confusion Matrix",
"Actual values", "Predicted values", n_image=0)
plot_confusion_matrix(fig, axs, local_conf_matrix,
"Local Perceptron Confusion Matrix",
"Actual values", "Predicted values", n_image=1)
Congrats !¶
You have figured out how to train your first Federated Sklearn classifier model !
If you want to practise more, you can try to deploy such classifier on two or more nodes. As you can see, Perceptron is a limited model: its generalization is SGDCLassifier, provided by Fed-BioMed as a FedSGDCLassifier Training Plan. You can thus try to apply SGDCLassifier, providing more feature such as different cost functions, regularizations and learning rate decays.
Don't miss out other tutorials about Federated Sklearn models, and consult user guide for further information.


