What's Fed-BioMed
Fed-BioMed, an open-source federated learning framework
What is Fed-BioMed?
Fed-BioMed is an open source project focused on empowering biomedical research using non-centralized approaches for statistical analysis. The project is currently based on Python and PyTorch, and enables developing and deploying federated learning analysis in real-world machine learning application.
The goal of Fed-BioMed is to provide a simplified and secured environment to:
- Easily deploy state-of-the art federated learning analysis frameworks,
- Provide a friendly user interface to share data for federated learning experiments,
- Allow researchers to easily deploy their models and analysis methods,
- Foster research and collaborations in federated learning.
Fed-BioMed is an ongoing initiative, and the code is available on GitLab.
What is Federated Learning ?
Introduction
Standard machine learning approaches require to have a centralized dataset in order to train a model. In certain scenarios like in the biomedical field, this is not straightforward due to several reasons like:
- Privacy concerns:
- General Data Protection Regulation (GDPR): General Data Protection Regulation (GDPR) – Official Legal Text
- Californian Consumer Privacy Act (CCPA): California Consumer Privacy Act (CCPA) | State of California - Department of Justice - Office of the Attorney General
- Health Insurance Portability and Accountability (HIPAA): Health Information Privacy regulation | U.S. Department of Health and Human Service | HHS official website
- Family Educational Rights and Privacy Act (FERPA): Family Educational Rights and Privacy Act (FERPA) | U.S. Department of Education official website
- Ethical committee approval
- Transferring data to a centralized location
This slows down research in healthcare and limits the generalization of certain models.
Federated Learning
Federated learning (FL) is a machine learning procedure whose goal is to train a model without having data centralized. The goal of FL is to train higher quality models by having access to more data than centralized approaches, as well as to keep data securely decentralized.
Infrastructure of a federated learning setting in healthcare
A common scenario of federated learning in healthcare is shown as follows:
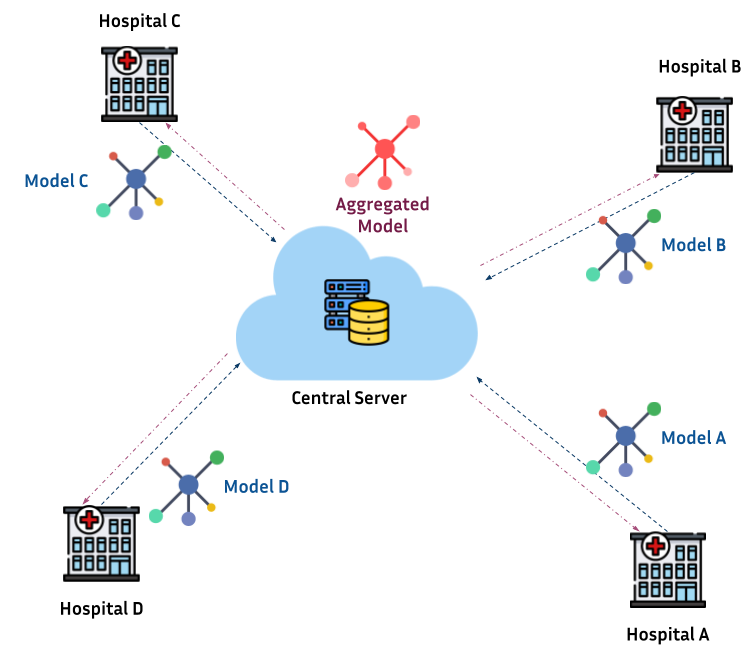
Hospitals (a.k.a. clients or nodes) across several geographical locations hold data of interest for a researcher. These data can be "made available" for local training but, only the model is authorized to be shared with a third thrusted party (e.g. research center). Once all the models are gathered, different techniques are proposed for aggregating them as a single global model. Then, the Aggregated model can be used as purposed (e.g. training a neural network for segmentation).
Theoretical background
One of the critical points in FL is knowing how to aggregate the models submitted by the clients. The main problem relies on finding the best set of parameters that define your model in function of the submissions made by the clients.
In a canonical form:
Where \(m\) is the total number of nodes, \(p_k>=0\), and \(\sum_k p_k=1\) , and \(F_k\) is the local objective function for the \(k\)-th node. The impact (contribution) of each node to the aggregation of the global model is given by \(p_k\).
One of the first proposed methodologies in FL for model aggregation was Federated Averaging FedAVG by (MacMahan et al, 2016), the idea behind it was to define the contribution of each node as \(p_k=\frac{n_k}{n}\) where \(n_k\) is the number of datapoints in the node \(k\) and \(n\) is the total number of observations studied.
Challenges in federated learning
The main challenges in FL are associated to:
-
Communication efficiency: number of iterations between nodes and central location to train an optimal model.
-
Data heterogeneity: how to build generalized models with heterogeneous data?
-
Security: adversarial attacks and data leakage.
References
-
Konečný, J., McMahan, et al. (2016). Federated learning: Strategies for improving communication efficiency. arXiv preprint arXiv:1610.05492.
-
Li, T., Sahu, et al. (2018). Federated optimization in heterogeneous networks. arXiv preprint arXiv:1812.06127.
-
Li, T., Sahu, A. K., Talwalkar, A., & Smith, V. (2020). Federated learning: Challenges, methods, and future directions. IEEE Signal Processing Magazine, 37(3), 50-60.


